-Search query
-Search result
Showing 1 - 50 of 627 items for (author: tai & l)

EMDB-35943: 
Cryo-EM structure of FFAR2 complex in apo state
Method: single particle / : Tai L, Li F, Sun X, Tang W, Wang J

PDB-8j23: 
Cryo-EM structure of FFAR2 complex in apo state
Method: single particle / : Tai L, Li F, Sun X, Tang W, Wang J

EMDB-16813: 
Tomogram of GBP1 coatomers assembled on brain polar lipid-derived small unilamellar vesicles.
Method: electron tomography / : Kuhm TI, Jakobi AJ
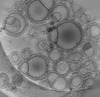
EMDB-16814: 
Tomogram of GBP1 coatomers assembled on brain polar lipid-derived small unilamellar vesicles.
Method: electron tomography / : Kuhm TI, Jakobi AJ
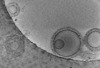
EMDB-16815: 
Tomogram of GBP1 coatomers assembled on brain polar lipid-derived small unilamellar vesicles.
Method: electron tomography / : Kuhm TI, Jakobi AJ

EMDB-41625: 
Type IV pilus from Pseudomonas PAO1 strain
Method: single particle / : Thongchol J, Zhang J, Zeng L

EMDB-41632: 
Asymmetric reconstruction of mature PP7 virions
Method: single particle / : Thongchol J, Zhang J, Zeng L

EMDB-41633: 
Type IV pilus from Pseudomonas PAO1 strain with PP7 Maturation protein
Method: single particle / : Thongchol J, Zhang J, Zeng L

EMDB-41675: 
The original consensus map of PP7 binding to T4P from PAO1
Method: single particle / : Thongchol J, Zhang J, Zeng L

EMDB-41780: 
Composite map of PP7 binding to type IV pilus from PAO1
Method: single particle / : Thongchol J, Zhang J, Zeng L

PDB-8tum: 
Type IV pilus from Pseudomonas PAO1 strain
Method: single particle / : Thongchol J, Zhang J, Zeng L

PDB-8tuw: 
Type IV pilus from Pseudomonas PAO1 strain with PP7 Maturation protein
Method: single particle / : Thongchol J, Zhang J, Zeng L

PDB-8tux: 
Capsid of mature PP7 virion with 3'end region of PP7 genomic RNA
Method: single particle / : Thongchol J, Zhang J, Zeng L

EMDB-16794: 
Cryo-EM structure of the human GBP1 dimer bound to GDP-AlF3
Method: single particle / : Kuhm TI, Jakobi AJ

EMDB-40687: 
PS3 F1 Rotorless, no ATP
Method: single particle / : Sobti M, Stewart AG

EMDB-40688: 
PS3 F1 Rotorless, low ATP
Method: single particle / : Sobti M, Stewart AG

EMDB-40689: 
PS3 F1 Rotorless, high ATP
Method: single particle / : Sobti M, Stewart AG

PDB-8spv: 
PS3 F1 Rotorless, no ATP
Method: single particle / : Sobti M, Stewart AG

PDB-8spw: 
PS3 F1 Rotorless, low ATP
Method: single particle / : Sobti M, Stewart AG

PDB-8spx: 
PS3 F1 Rotorless, high ATP
Method: single particle / : Sobti M, Stewart AG

EMDB-35940: 
Cryo-EM structure of FFAR3 bound with valeric acid and AR420626
Method: single particle / : Tai L, Li F, Sun X, Tang W, Wang J

EMDB-35941: 
Cryo-EM structure of FFAR3 complex bound with butyrate acid
Method: single particle / : Tai L, Li F, Sun X, Tang W, Wang J

EMDB-35942: 
Cryo-EM structure of FFAR2 complex bound with TUG-1375
Method: single particle / : Tai L, Li F, Sun X, Tang W, Wang J

EMDB-35944: 
Cryo-EM structure of FFAR2 complex bound with acetic acid
Method: single particle / : Tai L, Li F, Tang W, Sun X, Wang J

PDB-8j20: 
Cryo-EM structure of FFAR3 bound with valeric acid and AR420626
Method: single particle / : Tai L, Li F, Sun X, Tang W, Wang J

PDB-8j21: 
Cryo-EM structure of FFAR3 complex bound with butyrate acid
Method: single particle / : Tai L, Li F, Sun X, Tang W, Wang J

PDB-8j22: 
Cryo-EM structure of FFAR2 complex bound with TUG-1375
Method: single particle / : Tai L, Li F, Sun X, Tang W, Wang J

PDB-8j24: 
Cryo-EM structure of FFAR2 complex bound with acetic acid
Method: single particle / : Tai L, Li F, Tang W, Sun X, Wang J

EMDB-35932: 
Local refined cryo-EM reconstruction of Omicron BA.5 RBD in complex with 8-9D Fab
Method: single particle / : Xiang Y, Li R

EMDB-35934: 
Cryo-EM reconstruction of SARS-CoV2 Omicron BA.5 spike in complex with 8-9D Fabs
Method: single particle / : Xiang Y, Li R

PDB-8j1t: 
Local refined cryo-EM structure of Omicron BA.5 RBD in complex with 8-9D Fab
Method: single particle / : Xiang Y, Li R

PDB-8j1v: 
Cryo-EM structure of SARS-CoV2 Omicron BA.5 spike in complex with 8-9D Fabs
Method: single particle / : Xiang Y, Li R

EMDB-41983: 
Structure of the phage immune evasion protein Gad1 bound to the Gabija GajAB complex
Method: single particle / : Antine SP, Johnson AG, Mooney SE, Mayer ML, Kranzsuch PJ

PDB-8u7i: 
Structure of the phage immune evasion protein Gad1 bound to the Gabija GajAB complex
Method: single particle / : Antine SP, Johnson AG, Mooney SE, Mayer ML, Kranzsuch PJ

EMDB-36636: 
Cryo-EM structure of SARS-CoV-1 2P spike protein in complex with W328-6A1 IgG
Method: single particle / : Zhu JY

EMDB-36647: 
Cryo-EM structure of SARS-CoV-1 2P spike protein in complex with W328-6E10 Fab
Method: single particle / : Zhu JY

EMDB-36648: 
Cryo-EM structure of SARS-CoV-1 2P spike protein protomer in complex with W328-6E10 (local refine)
Method: single particle / : Zhu JY

EMDB-34550: 
Immune complex of W322-3D10 IgG binding the RBD of SARS-CoV-1 2P spike protein
Method: single particle / : Zhu JY, Zhou BN

EMDB-34553: 
Immune complex of W328-5B1 IgG binding the RBD of SARS-CoV-1 2P spike protein
Method: single particle / : Zhu JY, Zhou BN

EMDB-34557: 
Immune complex of W322-3E12 IgG binding the RBD of SARS-CoV-1 2P spike protein
Method: single particle / : Zhu JY, Zhou BN

EMDB-34558: 
Immune complex of W305-2A5 IgG binding the RBD of SARS-CoV-1 2P spike protein
Method: single particle / : Zhu JY, Zhou BN

EMDB-34559: 
Immune complex of W313-6D11 IgG binding the RBD of SARS-CoV-1 2P spike protein
Method: single particle / : Zhu JY, Zhou BN

EMDB-34560: 
Immune complex of W322-3B1 Fab binding the RBD of SARS-CoV-1 2P spike protein
Method: single particle / : Zhu JY, Zhou BN

EMDB-34561: 
Immune complex of W322-3G5 IgG binding the RBD of SARS-CoV-1 2P spike protein
Method: single particle / : Zhu JY, Zhou BN

EMDB-34562: 
Immune complex of W328-6F1 IgG binding the RBD of SARS-CoV-1 2P spike protein
Method: single particle / : Zhu JY, Zhou BN

EMDB-34599: 
Immune complex of W305-2G7 IgG binding the RBD of SARS-CoV-1 2P spike protein
Method: single particle / : Zhu JY, Zhou BN

EMDB-34600: 
Immune complex of W305-2A4 Fab binding the RBD of SARS-CoV-1 2P spike protein
Method: single particle / : Zhu JY, Zhou BN

EMDB-34601: 
Immune complex of W328-6G6 Fab binding the RBD of SARS-CoV-1 2P spike protein
Method: single particle / : Zhu JY, Zhou BN

EMDB-34604: 
Immune complex of W328-6H2 Fab binding the RBD of SARS-CoV-1 2P spike protein
Method: single particle / : Zhu JY, Zhou BN

EMDB-34605: 
Immune complex of W322-3F4 Fab binding the RBD of SARS-CoV-1 2P spike protein
Method: single particle / : Zhu JY, Zhou BN
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
